Lin Lab of RNA Modifications and Transcriptome Engineering
The Lin Lab studies RNA modifications (“epitranscriptomics”) in human health and disease. Post-transcriptional RNA processing and modifications are key mechanisms for gene regulation and functional diversity in eukaryotic cells. We develop and apply high-throughput sequencing strategies and transcriptome engineering technologies to study the regulation and function of RNA modifications, including alternative splicing, A-to-I RNA editing, and m6A RNA methylation.
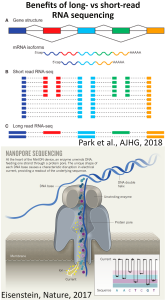 Transcriptome analysis using long-read nanopore sequencing
Transcriptome analysis using long-read nanopore sequencing
Long-read sequencing technologies, using third-generation DNA sequencers from Pacific Biosciences and Oxford Nanopore Technologies, are revolutionizing genomic research. These technologies have exciting transcriptomic applications, allowing direct resolution of transcript isoform structures and interrogation of repetitive RNA sequences. Using state-of-the-art nanopore long-read sequencing devices (MinION and GridION), we are developing new experimental methods to discover and quantify diverse RNA species in bulk tissues and single cells. By comparing the repertoire of full-length RNA transcripts between normal and diseased states (e.g. tumors), we hope to discover molecular markers or therapeutic targets of disease.
Functional consequences of A-to-I RNA editing events in UTRs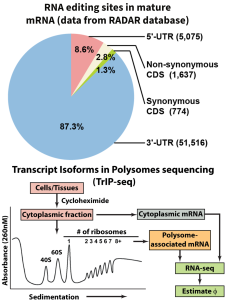
A critical, widespread mechanism for generating transcriptome diversity in eukaryotic cells, aberrations in RNA editing have been implicated in various diseases. Researchers have primarily studied the most abundant RNA editing type (A-to-I RNA editing) in protein-coding RNA regions, due to the potential for amino acid re-coding. However, based on an analysis of the RADAR database of A-to-I RNA editing events, nearly all such events (~96%) are located in untranslated regions (UTRs) of the mature mRNA. Moreover, their functions are largely unknown. By combining genomic, bioinformatic, and molecular approaches, such as Transcript Isoforms in Polysome sequencing (TrIP-seq), our lab studies the regulatory functions of A-to-I RNA editing events in 5’ and 3’ UTRs, as well as the roles of RNA editing in shaping complex traits and diseases.
Regulation of N6-methyladenosine by RNA binding proteins
N6-methyladenosine (m6A) is an abundant and dynamically regulated class of RNA base modification in mRNAs and non-coding RNAs. It affects multiple aspects of RNA metabolism and controls developmental transitions by regulating mRNA decay and translation. We are developing sensitive sequencing methods to detect RNA m6A methylation in a wide array of clinical and biological samples. Additionally, our lab uses transcriptome engineering technologies to investigate the regulatory and functional consequences of m6A methylation. For instance, we use the CRISPRi system to systematically knockdown individual RNA binding proteins (RBPs) and investigate the relationship between the RNA m6A methylome and the RBP-RNA interactome.



